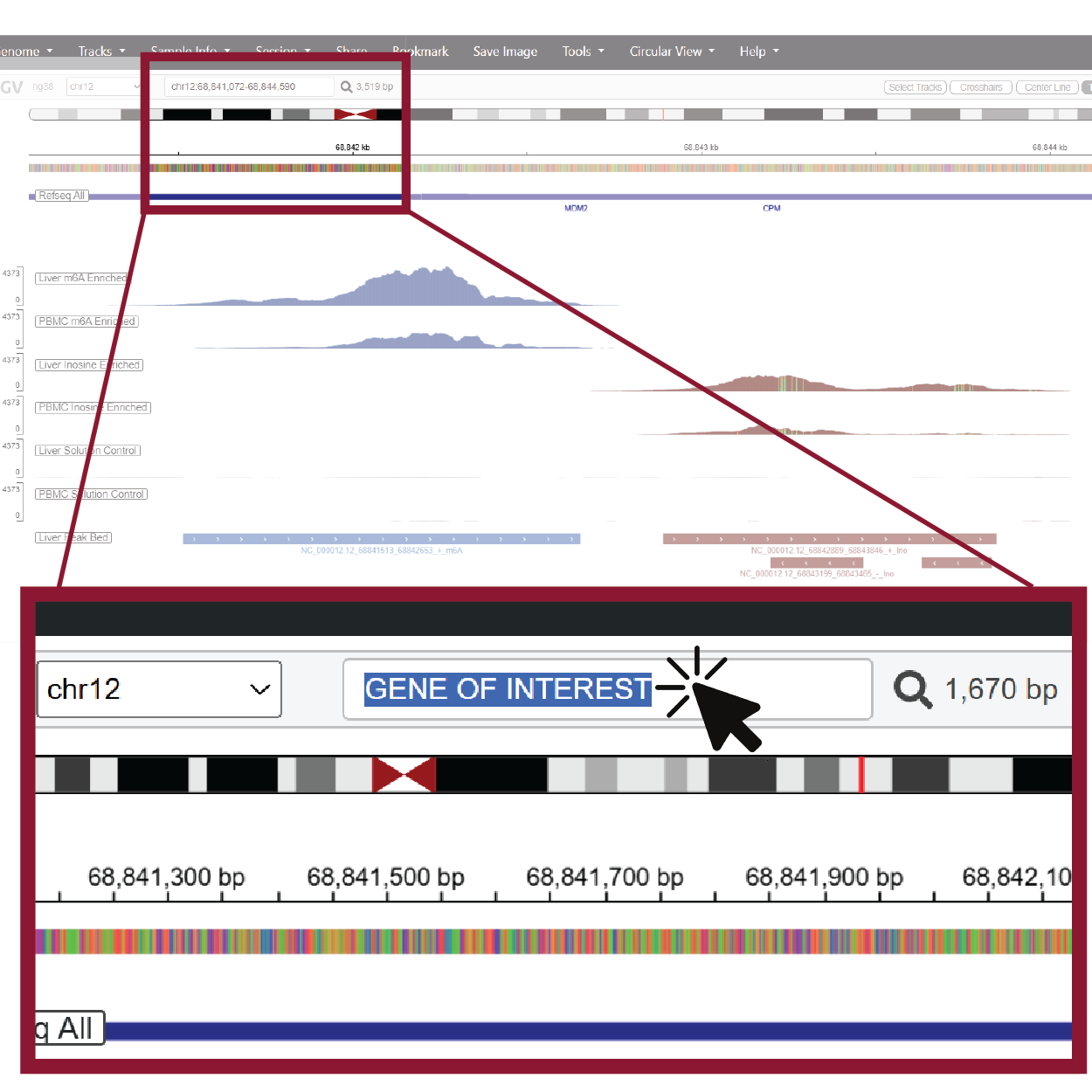
1. Use the search bar to navigate to other genes
The IGV search bar accepts gene names or genomic coordinates. Type a gene symbol or region into the box and press Enter, and the browser will jump directly to your region of interest.
Evening and midnight collections capture the Arabidopsis clock network at two late-day checkpoints, spanning wild type and single-gene knockouts in PRR and MYB/RVE clock modules.
| Field | Value |
|---|---|
| Organism | Arabidopsis thaliana (Col-0 background) |
| Collection times | CT20 (evening), CT24 (midnight) |
| Genotypes sampled | Col (WT), prr9, rve4, cca1, prr7 |
| Sample IDs (CT20) | Col_CT20, prr9_CT20, rve4_CT20 |
| Sample IDs (CT24) | cca1_CT24, prr7_CT24 |
CT = collection time (CT00 = midnight).
Samples and data provided by the Michael lab at Salk Institute.
The IGV sessions provide interactive visualization of m6A RNA methylation and RNA expression data comparing wild type (WT) against each knockout sample. Each session includes m6A-enriched tracks (showing modification sites) and control RNASeq tracks (showing expression levels), along with peak call annotations. By default, each session opens to TOC1, a gene critical for the function of the circadian clock. You can navigate to other clock genes (such as CCA1, PRR7, PRR9, or RVE4) or any region of interest using the search bar.

The IGV search bar accepts gene names or genomic coordinates. Type a gene symbol or region into the box and press Enter, and the browser will jump directly to your region of interest.
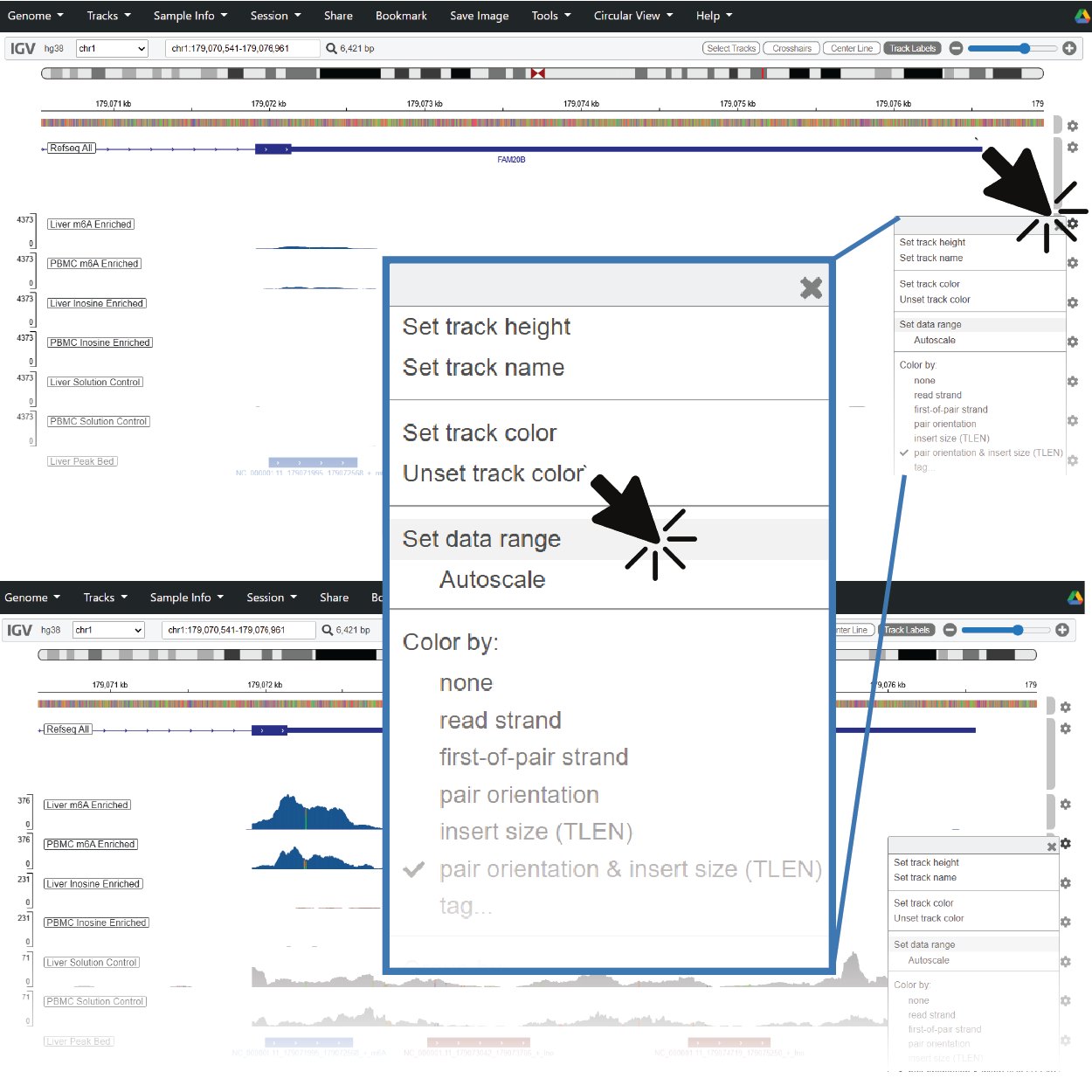
When you jump to a new region, coverage and modification tracks can span a wide dynamic range. If you don't see clear pileups or peak shapes, try right-clicking on the track to adjust the data range or enable autoscaling.
Plants do not just respond to light and temperature in the moment. They also anticipate daily change. The circadian clock is an internal timing system that helps a plant align biology with the day–night cycle, so processes like metabolism, growth, and stress responses occur at appropriate times of day.
In Arabidopsis, this timing system is built from a set of clock genes that regulate one another in feedback loops. Together, they generate rhythmic activity that repeats about every 24 hours, even under constant conditions. One common way to visualize these rhythms is to fuse a clock-controlled promoter to a luciferase (LUC) reporter. As clock-regulated transcription rises and falls, luminescence oscillates in a regular wave over time (Panel A).
Importantly, the circadian system is not controlled only at the level of DNA and transcription. The cell also tunes clock behavior after an mRNA is made, through processes such as RNA processing and chemical modifications. One of the best-studied RNA marks is m6A RNA methylation, which is deposited by a dedicated writer complex and is enriched on transcripts from clock-regulated and clock-associated genes. In plants, m6A can help connect environmental light signals to clock regulation, for example through light-sensing cryptochromes that recruit m6A machinery to influence clock transcripts.
When individual clock genes are disrupted, the oscillator can keep time differently. Some mutations make the rhythm run faster (short period), some make it run slower (long period), and some weaken rhythmicity altogether (arrhythmic) (Panel B). These oscillator changes matter because the clock helps coordinate downstream "output" traits, including seedling growth (often measured as hypocotyl length) and seasonal development such as photoperiodic flowering (Panel C). Changes to RNA methylation and RNA processing can also shift clock timing, and reduced m6A activity has been associated with longer circadian periods in Arabidopsis.
This demo dataset provides late-day snapshots (CT20 and CT24) from wild type and selected clock-gene knockouts. It is designed to illustrate how perturbing the clock can be reflected in molecular readouts, including RNA-state and epitranscriptomic signatures, at defined times of day.

Monitoring plant clock function and its phenotypic outputs. (A) After entrainment in light/dark cycles, luciferase (LUC) reporter rhythms are recorded in constant light. (B) Clock perturbations produce short-period, long-period, or arrhythmic oscillations; lowercase gene names indicate loss-of-function (knockout) and uppercase indicate constitutive overexpression. (C) These oscillator changes can translate into output phenotypes such as hypocotyl length and photoperiodic flowering time.
Figure adapted from Nagel, D.H.; Kay, S.A. Complexity in the Wiring and Regulation of Plant Circadian Networks. Current Biology 22 (2012) R648–R657. (Corrected figure and erratum: Current Biology 23 (2013) 95–96.) doi:10.1016/j.cub.2012.12.016
CIRCADIAN CLOCK ASSOCIATED 1
CCA1 is a MYB-domain transcription factor embedded in the Arabidopsis core oscillator. In the canonical feedback architecture, CCA1 (with LHY) represses evening-phased components such as TOC1 via promoter binding.
PSEUDO-RESPONSE REGULATOR 7
PRR7 participates in a morning-phased regulatory circuit: CCA1/LHY promote PRR7 expression, while PRR7 contributes repressive regulation that helps shape morning factor expression.
PSEUDO-RESPONSE REGULATOR 9
PRR9 is part of the morning loop promoted by CCA1/LHY. The review describes PRR-family regulation as transcriptional repression that helps coregulate the morning oscillator factors.
REVEILLE 4
RVE4 is a REVEILLE-family clock-linked regulator. RVE4 is a paralog of RVE6 and RVE8 which act to modulate the circadian period.